Year of publication
Contents
Year of publication#
Primary research questions:#
The notebook partly answers the following research question:
How is sharing affected by FOSS, Covid-19, publication type and year of publication?
Specifically we look at the number of shared models split into general health and Covid-19 subgroups. We also analyse how the number of shared models as a proportion of the literature.
1. Imports#
1.1. Standard Imports#
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
# for converting svg to tif
from PIL import Image
# set up plot style as ggplot
plt.style.use('ggplot')
1.2 Imports from preprocessing module#
# function for loading full dataset
from preprocessing import load_clean_dataset
2. Constants#
FILE_NAME = 'https://raw.githubusercontent.com/TomMonks/' \
+ 'des_sharing_lit_review/main/data/share_sim_data_extract.zip'
RG_LABEL = 'reporting_guidelines_mention'
NONE = 'None'
WIDTH = 0.5
3. Functions#
3.1. Functions to create summary statistics#
Two functions are used together in order to generate the high level results by year.
high_level_metrics- takes a subgroup of the dataset and generates summary statistics and countsanalysis_by_year- loop through the years passing each tohigh_levle_metricsand concatenates datasets at the end.
def high_level_metrics(df, name='None'):
'''A simple high level summary of the review.
Returns a dict containing simple high level counts
and percentages in the data#
Params:
-------
df: pd.DataFrame
A cleaned dataset. Could be overall or subgroups/categories
Returns:
--------
dict
'''
results = {}
included = df[df['study_included'] == 1]
available = included[included['model_code_available'] == 1]
results['n_included'] = len(included[included['study_included'] == 1])
results['n_foss'] = len(included[included['foss_sim'] == '1'])
results['n_covid'] = len(included[included['covid'] == 1])
results['n_avail'] = len(included[included['model_code_available'] == 1])
results['n_foss_avail'] = len(available[available['foss_sim'] == '1'])
results['n_covid_avail'] = len(available[available['covid'] == 1])
results['per_foss'] = results['n_foss'] / results['n_included']
results['per_covid'] = results['n_covid'] / results['n_included']
results['per_avail'] = results['n_avail'] / results['n_included']
results['per_foss_avail'] = results['n_foss_avail'] / results['n_foss']
# fix for 2019 as `n_covid` = 0
if results['n_covid'] > 0:
results['per_covid_avail'] = \
results['n_covid_avail'] / results['n_covid']
else:
results['n_covid_avail'] = 0.0
results['reporting_guide'] = \
len(included[included['reporting_guidelines_mention'] != 'None'])
results['per_reporting_guide'] = \
results['reporting_guide'] / results['n_included']
return pd.Series(results, name=name)
def analysis_by_year(df_clean, decimals=4):
'''
Conducts a high level analysis by year of publication
2019-2022
Params:
-------
df_clean: pd.DataFrame
Assumes a cleaned version of the dataset.
Returns:
-------
pd.DataFrame
Containing the result summary
'''
overall_results = high_level_metrics(df_clean, 'overall')
year_results = []
years = df_clean['pub_yr'].unique().tolist()
for year in years:
subset = df_clean[df_clean['pub_yr'] == year]
year_results.append(high_level_metrics(subset, name=str(year)))
year_results = [overall_results] + year_results
year_results = pd.DataFrame(year_results).T.round(decimals)
return year_results[sorted(year_results.columns.tolist())]
3.2 Functions to plot results#
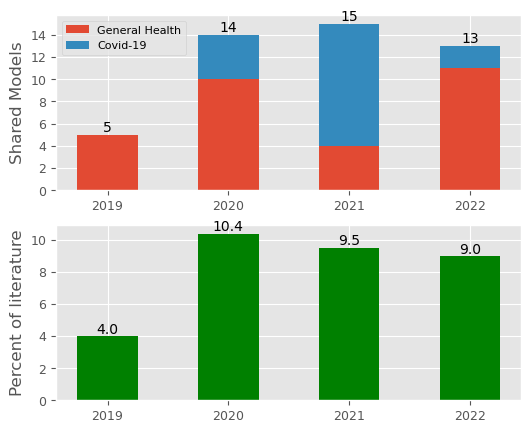
get_subgroups_as_dataframes. Creates simpler to use DataFrame containing annual summaries of shared and non-shared models. These are split into non-covid and covid subgroups.plot_sharing_by_year- plot with 2 subfigures. The top subfigure is a stacked barchart of general health models + covid-19 models that have been shared by year of publication. The lower subfigure presents yearly results as a percentage of total papers included in the study.
def get_subgroups_as_dataframes(yr_summary):
'''
Create seperate subgroups from the yr_summary table
and return as independent DataFrame objects. The overall column
is dropped and only annual figures are included.
Makes assumptions about the naming of colunms in yr_summary.
1. Non covid models shared
2. Covid models shared
3. Covid models not shared
4. Non covid models shared.
Params:
-------
yr_summary: pd.DataFrame
Dataframe containing summary of metrics by publication year
Returns:
-------
tuple
non-covid shared, covid shared, covid not shared, non covid not shared.
'''
# non covid shared models
non_covid_shared = yr_summary.T['n_avail'] \
- yr_summary.T['n_covid_avail']
# covid shared models
covid_shared = yr_summary.T['n_covid_avail']
# Covid models not shared
covid_not_shared = yr_summary.T['n_covid'] \
- yr_summary.T['n_covid_avail']
# non covid not shared
non_covid_not_shared = yr_summary.T['n_included'] \
- yr_summary.T['n_avail'] - yr_summary.T['n_covid']
# trim the overall column
return non_covid_shared[:-1], covid_shared[:-1], covid_not_shared[:-1], \
non_covid_not_shared[:-1]
def plot_sharing_by_year(n_general_models, n_covid_models,
per_models, xlabels, width=WIDTH, grid=True,
figsize=(6, 5)):
'''
Plot the number of models shared by year of publication (in citation).
In this function we split models into two subgroups: covid and non-covid
Non-covid models are refered to as 'general health models'
The function creates a figure with two subplots.
Subplot 1: A stacked barchart of general health models (non covid)
+ covid-19 models that have been shared by year of publication. A legend is
provided to distinguish between general and covid models.
Subplot 2: A bar chart expressing the total number of shared models as a
percentage of the number of included studies.
Params:
-------
n_general_models: pd.DataFrame
Count of non covid-19 models that have been shared by year of pub.
n_covid_models: pd.DataFrame
Count of covid-19 models that have been shared by year of pub.
per_models: pd.DataFrame
Percent of total studies included that have been shared by year of pub.
xlabels: list:
The years that represent the x-axis
width: float, optional (default=WIDTH)
A parameter to manipulated the width of the bars (applies to both plots)
grid: bool, optional (default=True)
Display grid lines (applies to both subplots)
figsize: tuple(int, int), optional (default=(6,5)
The size of the figure.
Returns:
--------
fig, ax
'''
fig, (ax1, ax2) = plt.subplots(nrows=2, sharex=False, figsize=figsize)
# include x, y grid
if grid:
ax1.grid(ls='--', axis='y')
ax2.grid(ls='--', axis='y')
# plot one: stacked bar chart. general health + covid models
y1 = ax1.bar(xlabels, n_general_models,
label='General Health', width=width)
y2 = ax1.bar(xlabels, n_covid_models,
label='Covid-19', bottom=n_general_models, width=width)
_ = ax1.set_ylabel('Shared Models')
_ = ax1.legend(ncols=1, loc='upper left', fontsize=8)
# force integer values on y axis
ax1.yaxis.set_major_locator(MaxNLocator(integer=True))
# include x, y grid
if grid:
ax1.grid(ls='--', axis='y')
ax2.grid(ls='--', axis='y')
ax1.bar_label(y2)
# set size of x, y ticks
ax1.tick_params(axis='both', labelsize=9)
ax2.tick_params(axis='both', labelsize=9)
# plot 2: percentage available by year
y2 = ax2.bar(xlabels, per_models * 100.0, label='All models',
width=width, color='green')
labels = [f'{m:.1f}' for m in (per_models * 100.0)]
ax2.bar_label(ax2.containers[0], labels=labels)
_ = ax2.set_ylabel('Percent of literature')
return fig, (ax1, ax2)
def plot_all_subgroups_by_year(non_covid_shared, covid_shared,
per_models, non_covid_not_shared,
covid_not_shared,
xlabels, width=WIDTH, grid=True,
figsize=(6, 6)):
'''
Plot the number of models across subgroups by year of publication (in citation).
In this function we split models into four subgroups: covid and non-covid
and shared and non-shared.
Non-covid models are refered to as 'general health models'
The function creates a figure with two subplots.
Subplot 1: A stacked barchart of unshared general health models (non covid)
+ unshared covid-19 + shared non-covid + shared covid that have been shared
by year of publication.
A legend is provided to distinguish between the four subgroups
Subplot 2: A bar chart expressing the total number of shared models as a
percentage of the number of included studies.
Params:
-------
non_covid_shared: pd.DataFrame
Count of non covid-19 models that have been shared by year of pub.
covid_shared: pd.DataFrame
Count of covid-19 models that have been shared by year of pub.
non_covid_not_shared: pd.DataFrame
Count of non covid-19 models that have NOT been shared by year of pub.
covid_not_shared: pd.DataFrame
Count of covid-19 models that have NOT been shared by year of pub.
per_models: pd.DataFrame
Percent of total studies included that have been shared by year of pub.
xlabels: list:
The years that represent the x-axis
width: float, optional (default=WIDTH)
A parameter to manipulated the width of the bars (applies to both plots)
grid: bool, optional (default=True)
Display grid lines (applies to both subplots)
figsize: tuple(int, int), optional (default=(6,5)
The size of the figure.
Returns:
--------
fig, ax
'''
fig, (ax1, ax2) = plt.subplots(nrows=2, sharex=True, figsize=figsize)
# include x, y grid
if grid:
ax1.grid(ls='--', axis='y')
ax2.grid(ls='--', axis='y')
# plot one: stacked bar chart. general health + covid models
y1 = ax1.bar(xlabels, non_covid_not_shared,
label='Closed General', width=width)
y2 = ax1.bar(xlabels, covid_not_shared,
label='Closed Covid-19', bottom=non_covid_not_shared,
width=width)
y3 = ax1.bar(xlabels, non_covid_shared,
label='Open General',
bottom=non_covid_not_shared+covid_not_shared, width=width)
y4 = ax1.bar(xlabels, covid_shared,
label='Open Covid-19',
bottom=non_covid_not_shared+covid_not_shared+
non_covid_shared, width=width)
_ = ax1.set_ylabel('Models')
_ = fig.legend(loc='upper left', ncol=2, bbox_to_anchor=(0.11, 1.0),
fontsize=9)
# include x, y grid
if grid:
ax1.grid(ls='--', axis='y')
ax2.grid(ls='--', axis='y')
ax1.bar_label(y4)
# set size of x, y ticks
ax1.tick_params(axis='both', labelsize=9)
ax2.tick_params(axis='both', labelsize=9)
# plot 2: percentage available by year
_ = ax2.bar(xlabels, per_models, label='All models',
width=width, color='green')
_ = ax2.set_ylabel('Percent of literature')
return fig, (ax1, ax2)
4. Read in data#
clean = load_clean_dataset(FILE_NAME)
5. Results#
5.1 Overall summary table#
# overall
year_summary = analysis_by_year(clean)
year_summary
| 2019 | 2020 | 2021 | 2022 | overall | |
|---|---|---|---|---|---|
| n_included | 126.0000 | 135.0000 | 158.0000 | 145.0000 | 564.0000 |
| n_foss | 18.0000 | 21.0000 | 35.0000 | 27.0000 | 101.0000 |
| n_covid | 0.0000 | 9.0000 | 35.0000 | 25.0000 | 69.0000 |
| n_avail | 5.0000 | 14.0000 | 15.0000 | 13.0000 | 47.0000 |
| n_foss_avail | 5.0000 | 7.0000 | 10.0000 | 7.0000 | 29.0000 |
| n_covid_avail | 0.0000 | 4.0000 | 11.0000 | 2.0000 | 17.0000 |
| per_foss | 0.1429 | 0.1556 | 0.2215 | 0.1862 | 0.1791 |
| per_covid | 0.0000 | 0.0667 | 0.2215 | 0.1724 | 0.1223 |
| per_avail | 0.0397 | 0.1037 | 0.0949 | 0.0897 | 0.0833 |
| per_foss_avail | 0.2778 | 0.3333 | 0.2857 | 0.2593 | 0.2871 |
| per_covid_avail | NaN | 0.4444 | 0.3143 | 0.0800 | 0.2464 |
| reporting_guide | 10.0000 | 21.0000 | 12.0000 | 29.0000 | 72.0000 |
| per_reporting_guide | 0.0794 | 0.1556 | 0.0759 | 0.2000 | 0.1277 |
5.4 All sharing subgroups by year of publication.#
fig, axs = plot_all_subgroups_by_year(non_covid_shared=non_covid_shared,
covid_shared=covid_shared,
per_models=year_summary.T['per_avail'][:-1],
non_covid_not_shared=non_covid_not_shared,
covid_not_shared=covid_not_shared,
xlabels=year_summary.columns[:-1],
grid=False)